The challenge of breeding for increased grain production in an era of global climate change and genomics

Summary
Grain from self-pollinating crops contributes more than 60% of annual global calories for human consumption, and demand is increasing.
Grain production in self-pollinating crops is extremely sensitive to the effects of heat and drought, and these stresses are increasing as a result of global climate change.
The rate of increase in grain yield of the world’s major self-pollinating crops is not keeping pace with demand and is threatened by global climate change.
Heat and drought stress tolerance are complex traits often present in wild or landrace relatives of crop plants. They are difficult to transfer by traditional breeding methods.
Genomic selection may help this process, but depends on high genetic diversity with associated molecular marker polymorphism in elite breeding populations.
The confluence of human population increase, climate change and genomics necessarily promotes change in the underlying methods of crop breeding.
The ‘animal model’ provides best linear unbiased prediction (BLUP) of breeding value based on information from all relatives in the pedigree across cycles of selection.
A plant version of the animal model is advocated for self-pollinating crops to increase genetic diversity in breeding populations and accelerate response to selection.
BLUP or genomic BLUP values can be estimated for drought and heat-stress tolerance, grain yield and quality and predictions are improved by integrating data across cycles of selection.
Genetic diversity is retained, the rate of inbreeding can be controlled and the potential for long-term genetic gain is increased. Pure lines will “spin off” from this rapid early generation crossing programme.
This change in crop breeding methods is motivated by the need to improve response to selection, including genomic selection, for grain yield in the face of global climate change.
Key words
crop breeding, genetic diversity, animal model, pedigree selection, genomic selection, optimum contribution selection, heat stress, drought stress
Glossary
Additive genetic variance: the heritable genetic variance in a population.
If selection occurs during the selfing process, additive genetic variance is lost from the population and cannot be restored when selection ceases.
Traditional breeding methods for self-pollinating crops cause a permanent loss of additive genetic variance when crossing occurs after selfing and selection of pure lines (see reference 12).
Animal model: a process of recording the phenotype and pedigree relationships in animal breeding (also used in perennial crop species), which provides best linear unbiased prediction (BLUP) of the breeding value of an individual based on all the phenotypic information available from itself and its relatives in the pedigree down to the base population.
The animal model has revolutionised commercial animal breeding over the past 30 years, with increased rate of genetic improvement while controlling the rate of population inbreeding through additional selection schemes such as optimal contribution selection.
BLUP: best linear unbiased prediction. In the animal model, the BLUP value is a selection index based on all the phenotypic information available from itself and its relatives, and is the predicted breeding value of the individual. Various software programs have been developed to calculate BLUP values in animal breeding programmes, and this software can be used in selfing crops.
F1 individuals: first filial generation (F1) individuals are the direct descendants of a cross. In animals, the parents are heterozygous and each F1 individual in a family is genetically unique (unless the sibs are identical twins). In traditional selfing crop breeding programmes, the parents are pure lines and F1 individuals from a cross are genetically identical. In this article, the animal model is applied to selfing plants, and the parents used for crossing are heterozygous and not pure lines, and their progeny are referred to as F1 individuals. Alternatively, “S0” can be used for the direct descendants of a cross between two heterozygous parents of a selfing crop species.
Genomic selection: a modification of the animal model, where, instead of pedigree information, a “realized” relationship matrix is constructed from whole-genome molecular marker data on the individual and its relatives in the pedigree tree. The genomic predicted breeding value is often referred to as G-BLUP. Potentially, this increases the accuracy of predicted breeding values.
Heterozygous: individuals with different forms of a gene from the mother and the father. F1 individuals are described as heterozygous for this gene.
Homozygous: individuals with only one form of a gene, either from the mother or the father. Pure lines are highly homozygous because they are the product of many generations of selfing.
Inbreeding: in selfing crops, inbreeding causes a loss of genetic diversity known as genetic drift, or the random loss of alleles due to small population size. Selfing crops do not suffer from “inbreeding depression” as observed in animals and cross-pollinating plants. This loss of genetic diversity limits long-term genetic gain in selfing crops. For more evidence on low effective population size and genetic drift in selfing crops, see references 8 and 9.
Optimum contribution selection: a selection method developed in the animal model which optimises genetic diversity and potential for long-term genetic gain in the breeding population. Individuals are selected for mating based on their BLUP or genomic BLUP value plus their potential to minimise inbreeding and contribute to long-term genetic gain. See reference 16.
Phenotype: the value of a trait measured on an individual. Phenotyping for heat or drought tolerance could occur on an F1 individual, or its self progeny, in the plant version of the animal model. Grain quality or grain yield must be measured on the self progeny of an F1 individual, in order to have enough seed to evaluate grain yield or quality in field plots.
Polymorphism: used in relation to molecular markers when they detect heterozygous molecular marker ‘genes’. Low rates of polymorphism reflect high rates of inbreeding.
Pure lines: these are formed when F1 individuals are selfed for several generations to produce near-homozygous lines that breed true.
Selfing crops: crops which produce grain by self-pollination. This includes most of the world’s most important grain-producing crops, such as wheat, barley, oats, soybean, millet, sorghum and canola. Selfing crops have traditionally been crossed after selection of pure lines, a process which may last several years and thereby slow genetic progress.
Self-pollination: this occurs when the male gamete (pollen) fertilizes the female gamete (ovule) on the same plant and produces seed from self-pollination. In cross-pollinating species such as lucerne (alfalfa), rye or turnip rape, seed production is the result of cross-pollination and self-pollination normally fails.
Introduction
More than 60% of calories for human consumption are produced annually by self-pollinating grain crops (1).
The 2009 Declaration of the World Summit on Food Security stated that a 70% increase in agricultural output was required by 2050 to keep pace with population increase, and that agriculture needed to adapt to climate change through a sustainable use of genetic resources (2).
Two serious threats to food production are the effects of heat and drought stress on selfing crops. New sources of heat and drought tolerance are often found in wild or landrace accessions stored in genetic resource centres (3, 4), but such traits are complex and specific strategies of crop breeding are necessary to exploit these valuable alleles in elite crop breeding programmes (5).
New breeding technologies, such as genomic selection, may help to increase food production (6, 15), but this is limited by the underlying breeding methods for selfing crops which are not sustainable in the long term (8).
A new crop breeding method, based on the animal model, was proposed recently for long-term and sustainable genetic improvement in selfing crops.
The method used all genetic relationships (with crossing and selfing in the pedigree) across cycles of selection, plus additive and non-additive genetic covariances between cycles, and accelerated response to selection for a low heritability trait (9). Genomic selection could add value to the process.
The confluence of several factors – increasing human population, increasing demand for calories from selfing crops, climate change leading to heat and drought stress, potential reductions in grain yield due to heat and drought stress, and new genomic technologies – necessarily stimulates change in the underlying breeding method for selfing crops.
Background
The traditional breeding method in selfing crops has been described as “selfing before crossing” (9).
This method developed naturally in selfing annual crops, where new cultivars are identified after selfing and selection of inbred lines, and are then used in crossing.
New cultivars of selfing crops “breed true” – farmers confidently retain seed knowing that seed harvested from a pure line is genetically identical to seed that was sown. Selfing and selection of superior homozygous pure lines is the ultimate goal of breeding in self-pollinating crops (10, 11).
However, this method of breeding selfing crops limits long-term and sustainable genetic gain (8). “Selfing before crossing” results in huge numbers of potential genotypes for testing and selection after each generation. Selection reduces the numbers of genotypes for testing during selfing, often with the aid of breeding technology (6).
Consequently, new pure line cultivars represent just a small fraction of the genetic variation available in each cross, and usually only a few elite lines are used as parents in the next round of crossing. These few elite parents with few ancestors in their pedigree, restrict the effective population size in selfing crops (8, 9).
In outcrossing species, such as animals or perennial trees, selection occurs on F1 individuals – where each sib from a cross is genetically unique and has a unique breeding value that can be predicted by phenotyping.
Only superior individuals are used in crossing, and these are identified on the basis of information from all relatives in the pedigree through a process that has become known as the ‘animal model’ (7). In such species, additive genetic variance in the population can be restored when selection is removed (12).
This is not the case with selfing crops, because additive genetic variance in the breeding population is reduced by selection during selfing, and this reduction in additive genetic variance is inherited by future generations (12, 13).
As a result, selfing crop breeding programmes tend to have lower genetic diversity than animal breeding programmes and longer generation intervals (8). This limits both short-term and longer-term responses to selection.
Elite crop breeding programmes also tend to become remote, genetically speaking, from wild and landrace accessions which may store many valuable genes (such as genes for heat and drought tolerance).
This increases the difficulty of migrating minor alleles for complex traits with low heritability from wild into elite breeding populations – and special breeding methods are needed to “bridge the gap” between wild or landrace accessions and elite crop breeding programmes (5, 14).
The reaction of most researchers and funders has been to seek technological solutions to these problems (6, 15). However, the underlying methods of breeding selfing crops should be critically examined first.
When the ‘animal model’ (7) was used in a selfing crop, this dramatically improved the rate of response of selection to a complex trait over two cycles of selection (9).
The method used pedigree and phenotypic information from relatives back to the base population. Such a method could be used to improve yield under heat and drought stress in elite crop breeding programmes.
The method will provide a sound basis for genomic selection in selfing crops, for the introduction of new alleles from wild and landrace relatives, and for the conservation of additive genetic variance for long-term genetic gain.
As with all breeding methods, appropriate selection methods are critical to minimise inbreeding. Optimal contribution selection was developed for long-term genetic gain in the animal model (16), but the software would need to be modified to include selfing in the pedigree. The animal model has a lot to offer breeding of selfing crops.
Using the animal model in selfing crop plants
The animal model uses information from relatives including ancestors, collateral relatives, and progeny to find the best linear unbiased prediction (BLUP) of breeding value for an individual (7).
BLUP is a selection index based on information from all relatives in the pedigree back to the base population, and has been very successful in breeding of animals and perennial tree crops where it has increased the rate of genetic gain without a large increase in inbreeding (8, 9).
In a typical animal pedigree (Fig. 1), the F1 offspring inherit half their genes from the dam and half from the sire, and because every parent is heterozygous, then every F1 sibling is heterozygous and genetically unique.
The relatives that contribute most to accuracy of BLUP values are parents, full-sibs and progeny, and the individual itself, but accuracy can be improved by including collateral relatives (full sibs, half-sibs), repeated records, and records from correlated traits (17).
A typical animal breeding programme has a database with names of individuals and records of phenotypic measurements on each individual at each date of measurement.
In addition, there is a record of the names of each parent for each individual (the pedigree records). These data grow over time, when new individuals are added at birth.
Most plant breeders have records which are similar to animal breeders, with both phenotypic and pedigree records on individuals. Many public or commercial databases are available to help plant breeders manage such records.
The entire data set in animal breeding, including many cycles back to the base population, are analysed by software programs such as ASReml-R (25) to solve linear mixed models including phenotypic and pedigree records across cycles of selection to arrive at the BLUP of breeding value for each individual.
The BLUP value is based on all records over all generations in the ancestry. This type of analysis over cycles of selection has not been attempted before in plant breeding until recently (9), where it was based on similar linear mixed models solved using ASReml-R software.
In traditional annual crop breeding programmes, cross progeny are selfed for several years after crossing (Fig. 2). This delays selection of suitable lines until superior inbred characteristics are identified, and slows selection cycles.
As indicated above, crossing after selection reduces additive genetic variance available for future genetic gain. These differences between typical animal and plant breeding programmes are represented by the sheep (F1 individuals) and canola (replicated F6 plots) in Fig. 3.
In a recent example in a selfing crop (9), only one generation of selfing occurred between each cross (9) (Figure 4).
The method was tested in field peas, Pisum sativum, the same species used by Mendel to demonstrate the nature of inheritance (9).
In annual selfing crop plants, the plant undergoes sexual reproduction during the growing season, normally before phenotypic records are obtained. Therefore it is not possible to phenotype, select and use F1 individuals for crossing before flowering.
In the method developed by Cowling et al. (9), F1 plants were assessed for resistance to a low-heritability trait, ascochyta blight disease (Didymella pinodes complex), during the growing season.
Self set seed was harvested from selected F1 plants and grown on to be used for crossing. Data were analysed over two cycles of selection, and analysis was improved by including genetic relationships and the additive and non-additive genetic covariances between cycles.
High accuracy of predicted breeding values (r > 0.8) was achieved on F1 plants, with >11% increase in disease resistance forecast in the next cycle based on 20% selection proportion (9).
Cowling et al. (9) incorporated both selfing and crossing in the pedigree of their ‘plant model’ version of the animal model (Fig. 4).
The plant model conformed to assumptions of the animal model with information combined across cycles of selection back to the base population, thereby avoiding the problem of bias in selection (18).
The key advantages of the method were shortened selection cycles compared with traditional methods (Fig. 4) and potentially high genetic diversity in the breeding population, because many heterozygous parents were used in crossing.
Another key benefit was the presence of many “augmented half-sibs” – self and cross progeny related by a common parent. Such relationships have never before been used in the animal model, and increased accuracy of prediction of breeding values in F1 individuals (9).
The method could be adapted to traits measured in harvested seed of F1 plants, such as grain yield or quality under heat and drought stress. However, to avoid high rates of population inbreeding, the selection of parents used in crossing should be optimised to retain genetic diversity and improve prospects for long-term genetic gain.
Such a method developed for the animal model is “optimum contribution selection” (16), and this method could be modified to include selfing in the pedigree of the plant model.
Genomic selection may increase accuracy of predictions of breeding value compared with BLUP selection as it has done in the animal model (19, 20).
This potential in plant breeding has stimulated many recent reviews, although the potential value is limited by population structure and lack of polymorphisms in breeding populations of selfing crops (21).
No current models of genomic selection in selfing crops integrate training of molecular markers on selection candidates across generations (22), although this is now possible in the plant version of the animal model (9).
The potential value of genomic selection should always be evaluated against BLUP selection based on pedigree relationship information (23).
Genomic BLUPs, based on phenotypic data and molecular genetic relationship information across generations, have been used to advantage in the animal model (19, 20).
The plant version of the animal model does not interfere with the selfing process in crop breeding, which can proceed as normal with the selection of pure lines from each generation (Fig. 5).
Crossing will occur with self progeny of superior F1 individuals, but normal procedures of selfing will occur on these parent plants to develop superior pure lines. Therefore, the plant version of the animal model requires only small modifications to existing breeding programmes.
The key change is to cross selected F1 individuals or their self progeny, based on their BLUP breeding values from analysis across cycles as in the animal model (9). For traits such as grain yield or quality, the phenotype must be measured on self progeny of F1 individuals.
Problems to be confronted
Research has begun on using the animal model to integrate data across cycles of selection in selfing crops (9), but much work remains to be done, for example, to modify the model for yield and grain quality measured on self seed of F1 individuals.
Research will be necessary to optimise selection based on BLUP values and genetic relationships, in order to minimise population inbreeding. It is not wise to rely on truncation selection as this will quickly narrow the population through inbreeding.
Developments in optimum contribution selection will be necessary to include selfing in the pedigree, which has not been done before (16).
Plant breeders will need to be familiar with software for spatial trial design such as DiGGer (24), to assemble pedigree information accurately on a database (including selfing), and to analyse field trial data and pedigrees across cycles of selection using software such as ASReml-R (25).
A full example of the model, including the phenotypic data, pedigrees with selfing and code for ASReml-R, is provided by Cowling et al. (9).
In this author’s experience, the cost of the method is not significantly different from previous annual budgets in crop breeding, and costs can be substantially reduced if doubled haploidy or genomic technology is not affordable in the programme.
This makes the method ideal for developing countries where plant breeding budgets are low, but demands for improvements from selfing crop breeding are high.
For existing high-cost breeding programmes, the method involves relatively minor modifications to existing procedures, but the change will be highly rewarded with increased genetic diversity and greater long-term genetic gain compared with current breeding methods for selfing crops.
Impact on policy
The method outlined here requires little additional funding and can be used by all countries (developed or developing) and structures of funding (public and private).
Some breeding programmes are funded entirely by public sources, and others are entirely funded privately. Most breeding programmes rely indirectly on publicly funded research, especially in the area of pre-breeding which develops new sources of genetic diversity.
All selfing crop breeding programmes are united by their dependence on breeding methods developed over 100 years ago. The new method outlined here is flexible and can be applied equally to pre-breeding programs and to elite commercial programmes.
The method was developed in animal breeding, and requires only minor adjustments to apply to selfing crop plants. The plant model advocated here differs in the timing of crossing to traditional breeding of selfing crops.
Crossing will occur on F1 individuals or their self progeny, selected for required traits, before extended selfing and selection of pure lines. This will improve long-term genetic gain and minimise loss of additive genetic variance through low effective population size.
Plant breeders and funders may ask: our breeding methods have worked for securely for over 100 years, so why change now? Why not continue selfing then crossing with elite pure lines? It is true that genomic selection or other molecular genetic technologies could help future advances in crop breeding (6, 15).
However, global food production and human survival in an era of rapid climate change demands critical examination of the underlying method of selfing crop breeding, which could exploit methods developed in the animal model to achieve better results in the medium and long-term.
Loss of additive genetic variance by selection during selfing before crossing (11) is crippling genetic advance in many breeding programmes, and limits our ability to improve complex traits such as heat and drought tolerance.
Genomic BLUPs may improve selection accuracy compared with pedigree BLUPs in selfing crops, but both genomic and pedigree methods of prediction require optimum contribution selection techniques to improve long-term genetic gain, as in animal breeding (22). Prediction of breeding value, by itself, is only one part of successful long-term genetic gain.
For pre-breeders, the method would be used to retain genetic diversity from a wide range of sources, such as landrace or wild types with heat and drought tolerance, while improving the population across cycles of selection with large effective population size.
Selection will focus on the trait(s) of interest, such as drought or heat tolerance, and large effective population size will reduce population inbreeding and loss of alleles by genetic drift. Pre-breeders should incorporate elite cultivars into their programmes, to keep the agronomic attributes relevant to elite commercial programmes in the target region.
For commercial breeders, the method provides an opportunity to achieve better results than competitors in the medium and long-term.
This applies equally to hybrid breeders as it does to breeders of selfing crops. Most hybrid breeders use selfing to produce superior inbred parent lines. The method outlined here allows migration of new genetic diversity and greater retention of existing genetic diversity – both of which set up the programme for long-term genetic gain in an era of changing climates.
A wise policy on the part of administrators and funders of crop breeding, both public and private, would be to compare traditional breeding alongside the new approach outlined in this article. Breeders normally do not require too much incentive to “try something new” if given support from their leaders.
Conclusions
A new breeding method based on the animal model is proposed for selfing crops, where crossing occurs before extended selfing and selection of inbred lines.
The method promises to improve medium to long-term genetic gain, with higher genetic diversity and more rapid response to selection.
The method will reduce the loss of additive genetic variance which happens when selfing and selection occurs before crossing of elite pure lines (12).
The procedure can be implemented readily with training in software programs for trial design (24) and analysis (25). Databases can be modified to include pedigree information on individuals and their relatives, as well as phenotypic data.
The main difference to the animal model, which animal breeders have been developing during the past 20-30 years of BLUP selection, is the inclusion of limited selfing in the pedigree tree (see reference 9). Therefore, most of the developments in the animal model are readily transferred to this new method in selfing crops.
The changes proposed here to existing crop breeding programmes (Fig. 5) are relatively small, but breeders need to see “proof of concept” to adopt new breeding methods and new technologies.
Proof of concept is now available for the new breeding method (9), and the next step is to encourage governments and private companies to invest in the training, computers and software to allow breeders to compete and achieve the best results in the medium to long-term.
Such changes have occurred in animal breeding over the past 30 years. If implemented now, the plant model will lead to significant improvements in 5-10 years, but these improvements will accumulate into the future to contribute to the 70% increase in agricultural output proposed by the 2009 Declaration of the World Summit on Food Security by 2050, with a sustainable use of genetic resources (2).
References
1. Nelson, G. C., M. W. Rosegrant, A. Palazzo, I. Gray, C. Ingerstoll, and R. Robertson, 2010 Food security, farming, and climate change to 2050: scenarios, results, policy options. Research Monograph. International Food Policy Research Institute, Washington, DC, USA.
2. Food and Agriculture Organisation (FAO) of the United Nations, Declaration of the World Summit on Food Security, Rome, 16–18 November 2009 (www.fao.org/wsfs/world-summit/en/).
3. Annisa, S. Chen, N. C. Turner and W. A. Cowling, 2013 Genetic variation for heat tolerance during the reproductive phase in Brassica rapa. J. Agron. Crop Sci. 199, 424-435.
4. Guo, Y. M., N. C. Turner, S. Chen, M. N. Nelson, K. H. M. Siddique and W. A. Cowling, 2014 Genotypic variation for tolerance to transient drought during the reproductive phase of Brassica rapa. J. Agron. Crop Sci. (in press)
5. Cowling, W.A., Buirchell, B.J., and D.E. Falk. 2009. A model for incorporating novel alleles from the primary gene pool into elite crop breeding programs while reselecting major genes for domestication or adaptation. Crop and Pasture Science 60:1009–1015.
6. Tester, M., and P. Langridge. 2010. Breeding technologies to increase crop production in a changing world. Science 327:818-822.
7. Lynch, M., and B. Walsh, 1998 Genetics and Analysis of Quantitative Traits, Sinauer Assoc Inc., Sunderland, USA.
8. Cowling, W.A. 2013. Sustainable plant breeding. Plant Breeding 132:1-9.
9. Cowling, W.A., Stefanova, K.T., Beeck, C.P., Nelson, M.N., Hargreaves, B.L.W., Sass, O., Gilmour, A.R. and K.H.M. Siddique. 2015. Using the animal model to accelerate response to selection in a self-pollinating crop. G3-Genes Genomes Genetics (in press).
10. Allard, R. W., 1999 Principles of Plant Breeding. Wiley, New York, USA.
11. Wricke, G., and W. E. Weber, 1986 Quantitative Genetics and Selection in Plant Breeding. Walter de Gruyter and Co., Berlin, Germany.
12. Cornish, M. A., 1990 Selection during a selfing programme. I. The effects of a single round of selection. Heredity 65: 201-211.
13. Cornish, M. A., 1990 election during a selfing programme. II. The effects of two or more rounds of selection. Heredity 65: 213-220.
14. Falk, D. E., 2010 Generating and maintaining diversity at the elite level in crop breeding. Genome 53: 982–991.
15. Varshney, R. K., K. C. Bansal, P. K. Aggarwal, S. K. Datta and P. Q. Craufurd, 2011: Agricultural biotechnology for crop improvement in a variable climate: hope or hype? Trends Plant Sci. 16, 363-371.
16. Henryon, M., T. Ostersen, B. Ask, A. C. Sørensen, and P. Berg, 2015 Most of the long-term genetic gain from optimum-contribution selection can be realised with restrictions imposed during optimisation. Genet. Sel. Evol. 47: 21
17. Simm, G., 1998 Genetic Improvement of Cattle and Sheep. Farming Press, Tonbridge, UK.
18. Piepho, H.-P., J. Möhring, A. E. Melchinger, and A. Büchse, 2008 BLUP for phenotypic selection in plant breeding and variety testing. Euphytica 161: 209–228.
19. Goddard, M. E., and B. J. Hayes, 2009 Mapping genes for complex traits in domestic animals and their use in breeding programmes. Nat. Rev. Genet. 10: 381-391.
20. Hayes, G. J., P. M. Visscher, and M. E. Goddard, 2009 Increased accuracy of artificial selection by using the realized relationship matrix. Genet. Res. 91: 47-60.
21. Desta, Z. A., and R. Ortiz, 2014 Genomic selection: genome-wide prediction in plant improvement. Trends Plant Sci. 19: 592-601.
22. Jonas, E., and D.-J. de Koning, 2013 Does genomic selection have a future in plant breeding? Trends in Biotechnol. 31: 497-504.
23. Henryon, M., P. Berg and A. C. Sørensen, 2014 Animal-breeding schemes using genomic information need breeding plans designed to maximise long-term genetic gains. Livestock Sci. 166: 38-47
24. Coombes, N. E., 2009 DiGGer, a spatial design program. Biometric bulletin, NSW Department of Primary Industries, Orange, NSW, Australia.
25. Butler, D. G., B. R. Cullis, A. R. Gilmour, and B. J. Gogel, 2009 ASReml-R reference manual. Version 3. Training Series QE02001, Queensland Department of Primary Industries and Fisheries and NSW NSW Department of Primary Industries. Available at: https://www.vsni.co.uk/resources/documentation/ (last accessed on 15 May, 2015).
Download pdf
Figures
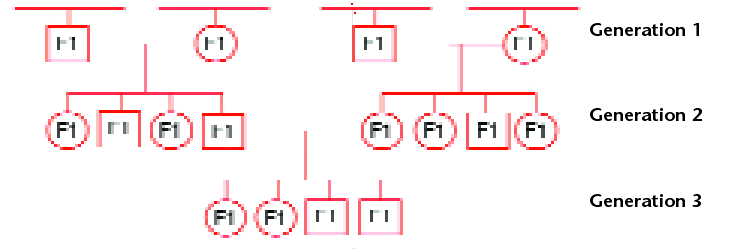
Fig 1. A typical pedigree in the animal model. Circles represent females and squares represent males. Every individual is an F1. In the animal model, data are analysed across cycles of selection using information from relatives in the pedigree. In each generation, F1’s with required traits are selected for crossing.
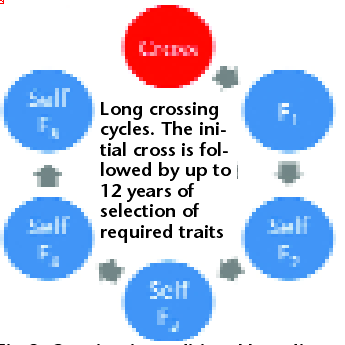
Fig 2. Crossing in traditional breeding programmes of self-pollinating annual crops is followed by several years of selfing. This leads to long cycle times (typically 6-12 years), low effective population size and a permanent loss of additive genetic variance in the population as a result of selection during selfing.

Fig 3. In animal breeding programmes, represented here by sheep in Australia, the phenotyping and selection is done on F1 individuals. In traditional selfing crop breeding programmes, represented by plots of canola lines in the rear of the picture, the phenotyping and selection is done during several generations of selfing, followed by yield and quality testing on F6-derived or later material.
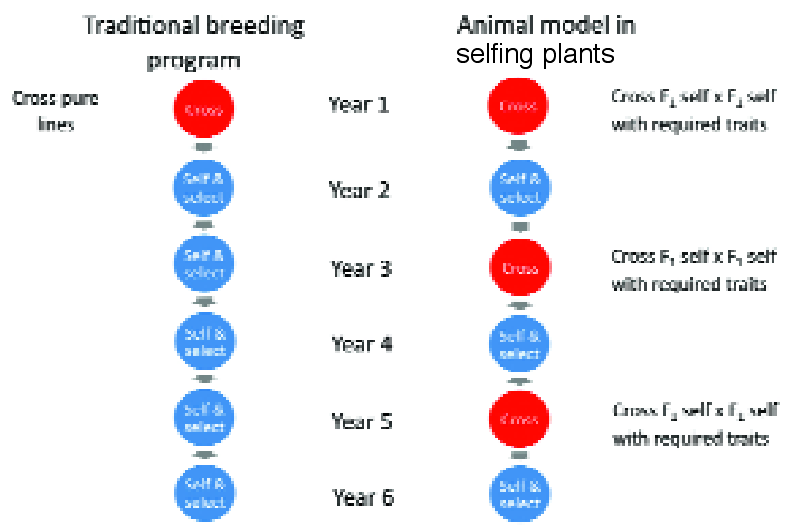
Fig 4. The cycle time in a traditional breeding programme of annual self- pollinating crops is shown on left taking at least 6 years, compared with the plant version of the animal model where the cycle time is just 2 years. The short cycle time on the right accelerates response to selection, and accuracy of prediction of breeding values is improved when the data are analysed across cycles according to the animal model (see reference 9).
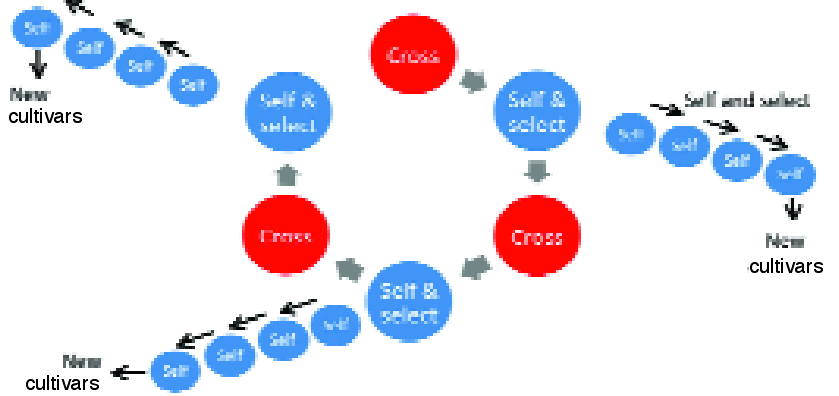
Fig 5. The plant version of the animal model allows selfing to continue on parent plants used in crossing (selfs of F1 individuals). The selfing programme continues “as normal” to produce and identify elite pure lines and cultivars, but crossing of the selfs of superior F1 individuals has already occurred several years earlier. This potentially accelerates response to selection and retains more genetic diversity and additive genetic variance for future generations.

